Novel brain MRI research may revolutionize study of neurological, psychiatric disorders
Two independent research papers published Oct. 10 in the journal Nature offer a glimpse into how genetic data from roughly 500,000 individuals in the U.K. could transform human health and disease research.
The data comes from the U.K. Biobank project—a prospective cohort study with genetic and phenotypic data collected between 2006 to 2010 from 500,000 individuals across the U.K. between the ages of 40 and 69 years. Biological measurements, lifestyle indicators, biomarkers in blood and urine, genome-wide genotype data and imaging of the body and the brain makeup the extremely large data set that is open to the public at no cost.
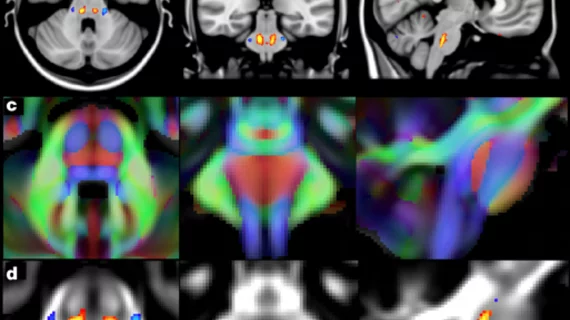
Of the two studies, one in particular focuses on brain-imaging data from more than 8,400 participants involved in the U.K. Biobank. The research led by Stephen Smith, PhD, from the Wellcome Center for Integrative Neuroimaging at the University of Oxford in the U.K., gives insight into the genetic makeup of the human brain relevant to neurological and psychiatric disorders, cognitive development and aging.
Having compared genetic data to 10,000 MRI brain scans, the research is the largest investigation of the genetic basis of brain structure and function, according to the researchers.
"We have found a genetic fingerprint on some of the most fundamental processes that allow us to think, act and function, from the size of the parts of the central nervous system that control sight, hearing, speech, emotions and actions to the integrity of the communications channels between them and the strength of the signals within them," Smith said in a prepared statement.
The MRI data was processed to generate a list of image-derived phenotypes (IDPs), or traits related to the brain structure and function identifiable through images.
Researchers studied the associations between IDPs and genetic variants and identified a total of 148 clusters of associations between single nucleotide polymorphisms and imaging phenotypes that replicate at certain levels.
“Notable significant, interpretable associations include: iron transport and storage genes, related to magnetic susceptibility of subcortical brain tissue; extracellular matrix and epidermal growth factor genes, associated with white matter micro-structure and lesions; genes that regulate mid-line axon development, associated with organization of the pontine crossing tract; and overall 17 genes involved in development, pathway signaling and plasticity,” Smith et al. wrote.
Overall, the study provides new data on the heritability of IDPs and may help improve the understanding of how genome-wide association studies on IDPs can be combined with genome-wide association study results on neurological and psychiatric disorders to learn more about the possible mechanisms of disease.
"The genetic basis of brain structure is largely unknown," co-author Gwenaëlle Douaud, PhD, said in a prepared statement. "We have found a very rich set of genetic effects which will help us understand better the mechanisms by which the brain develops, functions, becomes damaged by disease and heals itself.”
The U.K. Biobank is funded mainly by the U.K. Medical Research Council and the Wellcome Trust medical charity in London.